URTIRISK
The software URTIRISK allows the user to observe the evolution of the allergic risk associated with the presence of pine processionary moth, over a year and throughout the French territory. This software is mainly based on the observations of the population range carried out by INRA URZF (UR633).
Data
The software is based on the following data:
1. Position of the colonization front (observed by INRA URZF): Northern front (2010-2011) and altitudinal front in the Massif Central (2005-2006).
2. IGN map showing the location of pine trees.
3. Chart describing the life cycle of the PPM and its variations between southern France and the colonization front, as a function of latitude. This chart was obtained by adapting the observations of Huchon and Démolin (1970) to the current geographical distribution of the PPM.
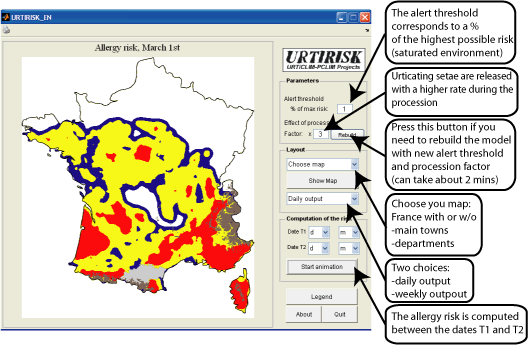
Computing the allergy risk
The allergy risk depends on the time of year, and, of course, on the presence of caterpillars.
The production of urticating setae begins during the larval stage "L3" (Huchon and Démolin 1970). We begin by defining a potential allergy risk R0 which depends on the latitude and on the time of year, but which is independent on the presence of PPM. The computation of this potential allergy risk is based on the following assumptions :
1. There is a time t0, which corresponds to the beginning of the larval stage L3 and a time t1 which corresponds to the beginning of the procession. These times depend on the latitude only. They are computed on the basis of the chart in Démolin and Huchon (1970), adapted to the current geographical distribution of the PPM.
2. The duration of the procession is fixed to 30 days.
3. Urticating setae are released with a constant rate between t0 and t1, and with another (higher) rate during the procession.
4. The half-life of the setae is fixed to 10 days in the current version of the software.
Then, we define a function R1, which depends on the position (latitude, longitude), and roughly describes the distribution of the PPM nests:
5. We build a map of the potential area of distribution of the PPM, using only the northern front and the altitudinal front in the Massif Central together with the altitudinal limit of 1400m.
6. Using the IGN map showing the location of pine trees, we restrict the area of distribution of the PPM to the locations where pine trees are present.
7. We use a smoothing kernel to take into account the uncertainty related with the presence of unlisted isolated host trees and PPM nests beyond the colonization front.
By overlaying maps R0 and R1, we obtain a model for the risk R as a function of position (latitude, longitude) and date.
Outputs
The risk R is normalized (in time and space), so that the maximum risk value is R=1. We define a warning threshold S in terms of the maximum risk. The URTIRISK software produces a map with four risk levels: no risk for R <S/10 (in white), low risk to S/10 ≤R <S (blue), medium risk for S ≤R <10S (in yellow) and very high risk for R≥ 10S (red).
Download
NEW (2024) : URTIRISK is available as Jupyter notebook
The Matlab® source code (for Matlab R2011a or later) of the URTIRISK software is available:
An executable version for Windows is also available:
ENGLISH executable version for Windows 32bits
ENGLISH executable version for Windows 64bits
Installation
If you already have Matlab® (R2011a or later) intalled on your computer:
1. Download the source code.
2. Unzip all the files of the urtirisk.zip archive in a folder (ex: in "matlab/myfolder").
3. Open Matlab® and seclect "matlab/myfolder" as you current folder.
4. Launch the URTIRISK.m file (or URTIRISK_EN.m file for the english version).
If Matlab® is not intalled on your computer:
1. Download the executable version.
2. Download and install the MATLAB compiler Runtime (MCR) corresponding to your OS.
3. Launch the URTIRISK.exe file. First execution can be lengthy because of MCR initialization.
Contact
Lionel Roques: lionel.roques@inrae.fr
